About Me
Paul Babitzke obtained his B.A. in Biomedical Science at St. Cloud State University and his Ph.D. in Genetics from the University of Georgia. He worked as a postdoctoral scientist for 3 years at Stanford University prior to joining the faculty at Penn State University in 1994. He is currently the Don Bryant Chair in Microbial Physiology and Director of the Center for RNA Molecular Biology.
Research Interest
Regulation of gene expression by RNA structure and RNA-binding proteins
Research Summary
Gene expression is regulated at the level of transcription, translation and mRNA stability. The transcription cycle consists of initiation, elongation and termination, each of which can be regulated. We are investigating fundamental mechanisms that regulate transcription elongation and termination. We also investigate a variety of transcription attenuation (regulated termination) and translation repression mechanisms in which binding of proteins (or small metabolites) to the nascent RNA dictates whether transcription terminates in the 5' leader region or continues into the downstream coding sequences.
The general transcription elongation factors NusA and NusG participate in attenuation mechanisms by stimulating RNA polymerase pausing at a region preceding the critical overlap between competing antiterminator and terminator structures. Pausing provides additional time for binding of the protein or metabolite to the nascent RNA. NusA stimulates pausing by assisting with pause hairpin formation, while NusG makes sequence-specific contacts with T residues in the non-template DNA (ntDNA) strand within the paused transcription bubble, thereby allosterically preventing nucleotide incorporation. Using a genomic approach called RNET-seq, we identified >1600 NusG-dependent pause sites in the B. subtilis transcriptome. Pausing at several of these sites were subsequently shown to regulate transcription termination in the 5' leader region.

Intrinsic (Rho-independent) and Rho-dependent termination are the two transcription termination mechanisms identified in bacteria. Canonical intrinsic terminators consist of a strong RNA hairpin followed by a U-rich tract. Term-seq studies in B. subtilis demonstrated that NusA, NusG, and Rho stimulate intrinsic termination. NusA-stimulates termination by assisting with terminator hairpin formation, NusG stimulates termination by stimulating pausing at the site of termination, while Rho stimulates termination by preventing the formation of inadvertent antiterminator-like structures. Of particular interest, intrinsic termination is also known as Rho-independent termination.
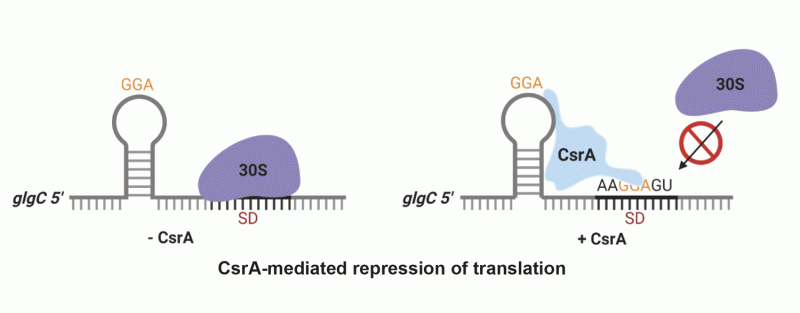
Another major effort in the lab involves genetic and biochemical characterization of the Csr global regulatory system. Csr regulates several hundred E. coli genes, thereby mediating global changes in cellular physiology during the transition between exponential and stationary phase growth. Four major components of Csr include an RNA binding protein (CsrA), two small RNA (sRNA) antagonists of CsrA (CsrB and CsrC), and CsrD, a protein that targets degradation of CsrB and CsrC by RNase E. CsrA represses translation initiation of numerous genes by binding to their translation initiation regions. Bound CsrA prevents ribosome binding, thereby repressing a variety of processes, including gluconeogenesis, glycogen biosynthesis, quorum sensing and biofilm formation. In contrast, CsrA activates glycolysis, acetate and iron metabolism, and motility. CsrA activates flagella biosynthesis by preventing degradation of the flhDC transcript by RNase E. We recently identified the first known CsrA-mediated translational activation mechanism. We are continuing to explore this global regulatory system in E. coli using omics, genetics, and biochemical approaches.
Honors and Awards
- Chair, NIGMS Microbial Physiology and Genetics-subcommittee 2 (MBC-2) (2004)
- Chair, Division H, American Society for Microbiology (ASM) (2006)
- Daniel R. Tershak Teaching Award (2009)
- Divisional Group IV Representative, American Society for Microbiology (ASM) (2011-2015)
- Charles E. Kaufman New Initiative Research Award (2016)
- Fellow, American Association for the Advancement of Science (AAAS) (2016)
- Fellow, American Academy of Microbiology (AAM) (2017)
- St. Cloud State University Biological Sciences Distinguished Alumni Award (2018)
- Sigma Xi, The Scientific Research Honor Society (2019)
- Stanley R. Person Professor of Molecular Biology endowed professorship (2021)
