
A tiny molecular structure that looks like a bubble may be able to significantly improve medical imaging, according to a Penn State research team. Called gas vesicles (GVs), these structures are naturally produced by certain microorganisms and are responsible for controlling the microorganism’s buoyancy in water. Researchers can genetically engineer human cells to produce these gas vesicles, resulting in an ultrasound contrast medium capable of revealing deep tissue structures at the resolution of a single cell.
The problem is that the process to engineer such cells is costly and arduous. To make the process easier, Lance Lian, associate professor of biomedical engineering and of biology at Penn State, led a team in developing a more efficient approach. They published their work in Bioengineering & Translational Medicine.
“The great thing about our approach is that it doesn't require the tedious and time-consuming single-cell cloning and sorting methods,” said Lian, co-corresponding author on the paper. “Instead, we can work with a mixture of cells and still get strong and reliable ultrasound contrast.”
In the context of this research, "single-cell cloning" refers to a labor-intensive process of isolating and cultivating individual cells that have undergone specific genetic modifications, such as incorporating the desired genes to produce GVs. Researchers typically do this to ensure that they are working with a population of cells that have uniform genetic characteristics.
To eliminate this complex step from the process, the team’s approach hinged on three specific DNA constructs, which, when integrated into the genetic material of cells, facilitate the expression of the genes responsible for creating GVs.
DNA constructs refer to engineered segments of DNA that carry specific genetic instructions. In this work, the researchers designed DNA constructs to act as genetic templates, guiding the cells to produce the components necessary for forming GVs.
The researchers also integrated a clock-like system called the "doxycycline TET-On 3G system" into these DNA constructs. It triggers the production of GVs within the cell at exactly the right time.
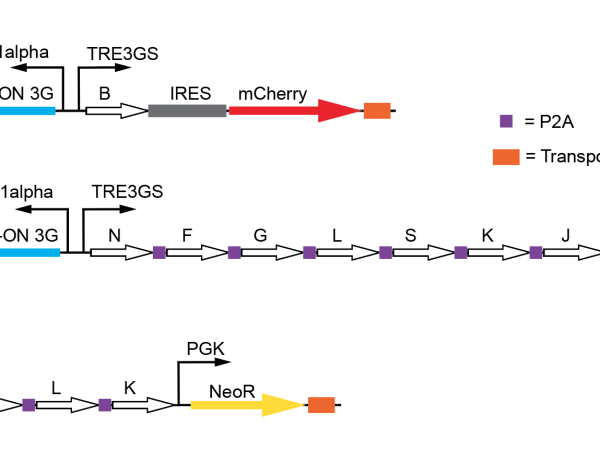
In addition to this precise timing mechanism, the team made custom modifications to the three DNA constructs by adding unique drug-resistant genes to each one. When researchers treated a cell population with the drugs, any cells that had not successfully incorporated all three DNA constructs died off. This method, referred to as "drug selecting," made it possible to isolate a uniform group of genetically modified cells that could produce GVs.
As a result, the prior demanding and intricate single-cell cloning process became unnecessary, Lian said.
“I think it's a classic example of when biology gets difficult, if you incorporate a drug resistant gene downstream of your gene of interest, it makes things easier,” said Alessandro Howells, a doctoral student in the Lian lab and the first author of the paper. He explained that by adding the drug-resistant genes, they essentially marked the cells capable of producing GVs.
Lian said he can envision, now that the process to produce them is more efficient, that GVs will have applications in therapeutic medical interventions in the future.
Scientists have been exploring ways to use GVs with ultrasound applications since they discovered it was discovered possible in 2014. Ultrasound uses sound waves to image biological tissue. When the sound waves hit something of a different density, such as bone, some of the sound bounces back and the ultrasound system can determine how deep that object is, building a picture of internal anatomy. This imaging method is non-invasive and does not expose the tissue to radiation.
According to Lian, GVs could someday replace the only currently clinically approved ultrasound agent, microbubbles. Smaller than a red blood cell, microbubbles are too large to use for molecular imaging. Given their much smaller size, Lian said, GVs could be used as a biological contrast agent for imaging deep into tissue, allowing researchers to non-invasively visualize structures to individual cells.
When GVs are hit with ultrasound waves, they alter their shapes. The signal that these buckling GVs make can be detected with the ultrasound system, and researchers can see exactly where those cells are. What's remarkable about the GV method is that it allows imaging of cells even when they're embedded several centimeters deep, according to Lian.
“Imagine if we can use this technology to monitor if transplanted therapeutic cells are still in the body,” Lian said, referring to cells engineered to replace or complement malfunctioning cells in the body, such as pancreatic beta cells his team developed to treating type 1 diabetes. “Sometimes they are destroyed by the body’s immune system, and we just have to wait and see if a treatment is not working. With GVs, we can continuously monitor the cells through ultrasound imaging and monitor a therapeutic intervention in real-time. I think that this technology will have enormous applications in biomedical research.”
The researchers made their constructs available through addgene, a nonprofit plasmid repository, enabling researchers from around the world to access their work.
In addition to Howells and Lian, other authors on the paper include Phoebe J. Welch, John Kim, Craig R. Forest and Chengzhi Shi, all with the Georgia Institute of Technology. Lian is also affiliated with the Penn State Huck Institutes of the Life Sciences.
The National Science Foundation supported this work.
