Precise control of gene expression—ensuring that cells make the correct components in the right amount and at the right time—is vital for all organisms to function properly. Cells must regulate how genes encoded in the sequence of DNA are made into RNA molecules that can carry out cellular functions on their own or be further processed into proteins. One way gene expression is regulated is by pausing “transcription”—the process by which RNA is synthesized from its DNA template by an enzyme called RNA polymerase. Now, researchers have worked out the mechanism of transcription pausing in some bacteria using cryo-electron microscopy (cryo-EM), which allows them to determine to atomic scale the structures of the RNA polymerase before, during, and just after a pause of RNA production. Elucidatiing the mechanism of pausing transcription is crucial to underestanding basic cellular function.
One of the key components of transcription pausing in the bacteria is a protein factor called NusG, which is conserved across organisms, including humans, such that the pausing mechanism revealed by this study may be broadly applicable for understanding gene regulation in all organisms on earth. The insight could also be used to identify new anti-bacterial agents that target and inhibit transcription pausing thus disrupting proper gene expression and cellular function.
A paper describing the research by a team of Penn State scientists appears online February 6, 2023 in the journal Proceedings of the National Academies of Science.
“To function properly, cells must precisely control gene expression to ensure that they are making the selected proteins and functional RNAs in the appropriate amount and at the appropriate time,” said Kastuhiko Murakami, professor of biochemistry and molecular biology at Penn State and one of the leaders of the research team. “Gene expression can be regulated in multiple ways even when RNA polymerase is actively making RNA such as pausing of transcription. Here, we used cryo-EM structure determination and structure-based biochemical assays to identify the interactions among RNA polymerase, DNA and NusG, as well as structural changes of RNA polymerase that take place when RNA polymerase pauses during transcription”.
RNA polymerase is a molecular machine composed of several functional domains. It also interacts with a number of other protein co-factors that help regulate the timing and amount of RNA produced. Work in the lab of Paul Babitzke, Stanley Person Professor of Molecular Biology at Penn State and a leader of the research team, had shown that one of those co-factors, the protein called NusG, was crucial for pausing transcription.
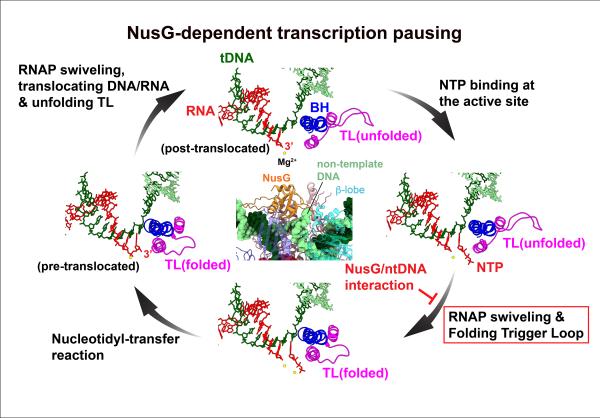
“We showed experimentally that NusG played an important role in pausing transcription by recognizing a specific short DNA sequence motif,” said Babitzke. “We found over 1600 of these sequence motifs across the genome of the bacteria, Bacillus subtilis, that are involved in NusG-dependent pausing of transcription. DNA is double stranded and only one strand—called the template strand—is transcribed into RNA. Interestingly the recognition motif for NusG is located on the non-template strand. To understand how NusG interacting with the non-template DNA could lead to pausing, we wanted to see what may be happening structurally to the complex. Cryo-EM has allowed us to do just that.”
The research team identified DNA and RNA sequences that cause transcription to pause in the presence of NusG, and then reconstituted the transcription complex using DNA, RNA and NusG together with RNA polymerase. The samples were frozen and then used for structural studies by cryo-EM using the state-of-the-art cryo-electron microscopy facility at the Penn State Huck Institutes of the Life Sciences.
In the structure of the paused transcription complex, the team revealed that NusG interacts with the non-template DNA strand by inserting it into a narrow cavity between NusG and an RNA polymerase domain called the beta lobe. But this interaction occurs far away from the active site of RNA polymerase, so how does it cause transcription to pause? The research team used unique information obtained by the cryo-EM structural study to address this question.
“When we freeze the sample, the reconstituted transcirption complexes can each be at slightly different stages in the transcription process,” said Rishi K. Vishwakarma, assistant research professor of biochemistry and molecular biology at Penn State and the first author of the paper. “With cryo-EM, we can use data from the single sample to determine a series of structures showing distinct stages of transcription pausing, then arrange these structures sequentially. We used them to reconstruct a movie of transcription complexes to observe what happens to the transcription complex when transcription pauses, sort of like a flipbook can represent movement from a series of still images.”
“It took some time to clarify how the interaction of NusG and the non-template DNA pauses transcription,” said Murakami. “One afternoon, when I was watching a movie of transcription complexes determined in this work, I noticed that a large conformational change of RNA polymerase—the swivel module rotation—is linked to trigger loop folding, an essential conformational change of RNA polymerase for RNA synthesis. When the non-template DNA is trapped between NusG and the RNA polymerase beta lobe, it prevents rotation of the swivel module, thereby interfering with trigger loop folding and RNA syntheis, sort of like getting something stuck in one of the gears of an engine that will impact other parts of the engine and stop the car. I was very excited about this finding and couldn’t stop watching the movie as my ideas solidified.”
Because transcription pausing is a temporary mechanism for stopping RNA synthesis, the team also looked at structural changes of the transcription complex as it escaped from pausing by preparing a sample containing the paused transcription complex and the RNA polymerase substrate (NTP) and determing cryo-EM structures. In the transcription complex, which had escaped from the pause, the non-template DNA was no longer trapped between NusG and the RNA polymerase beta lobe, which allowed the swivel module to once again rotate such that the trigger loop could fold properly.
“Because most of the components of this process are conserved from bacteria to humans, we are interested in knowing if the mechanism is also conserved,” said Murakami. “We are working with other cellular RNA polymerases such as from archaea and eukaryotes. We were fortunate to obtain a eukaryotic RNA polymerase isolated from Drosophila (fruit flies) from David Gilmour at Penn State, who has been studying transcription pausing in eukaryotes for decades. We will determine how paused RNA polymerase structures look in archaea and eukaryotic organisms.”
In addition to Murakami, Babitzke and, Vishwakarma, the research team at Penn State includes M. Zuhaib Qayyum, who is now a scientist at St. Jude Children's Research Hospital in Memphis, TN. The research was funded by the U.S. National Institutes of Health.
